The original nature of DNA replication
Background
Recombination-driven replication (RDR) likely represents an ancient strategy for initiating genomic replication and many questions remain regarding how the use of RDR versus origin-dependent replication (ODR) is regulated.
- Determine how environmental conditions impact the use of RDR versus ODR in T. kodakarensis. Substantial preliminary data supports the hypothesis that at reduced temperatures (65˚C) ODR dominates, whereas as optimal conditions (85˚C) RDR appears to serve as the major mechanism of DNA replication. Three well-conserved and often essential replication factors in T. kodakarensis are encoded with inteins, and we will detail temperature-dependent effects on protein splicing for Pol B, RadA, and MCM3.
- Establish the genetic requirement for sequences and factors directing ODR and RDR.
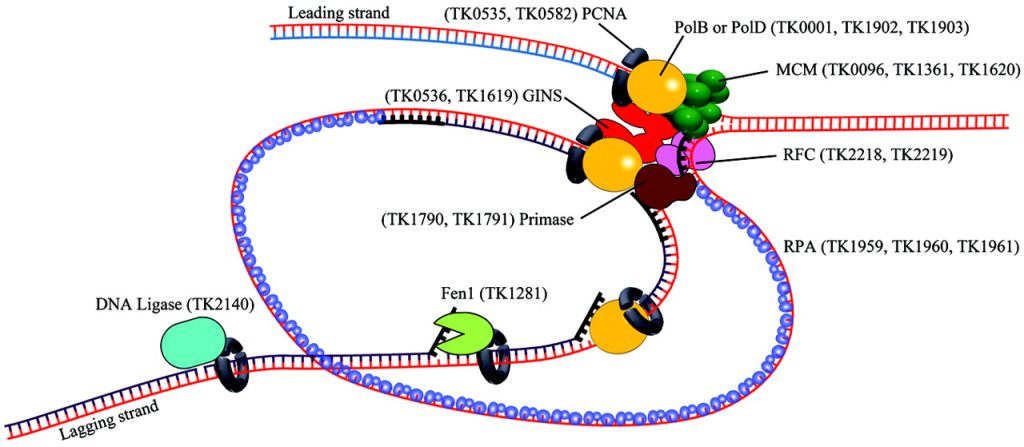
- Define the replisome components for ODR and RDR.
RELEVANT PUBLICATIONS
- Gehring AM, Astling DP, Matsumi R, Burkhart BW, Kelman Z, Reeve JN, Jones KL, Santangelo TJ. Genome Replication in Thermococcus kodakarensis Independent of Cdc6 and an Origin of Replication. Front Microbiol. 2017 Oct 27;8:2084.
- Cubonova L, Richardson T, Burkhart BW, Kelman Z, Connolly BA, Reeve JN, Santangelo TJ. Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J Bacteriol. 2013. May; 195(10):2322-8.
- Liman GLS, Stettler ME, Santangelo TJ. Transformation techniques for the anaerobic hyperthermophile Thermococcus kodakarensis. Methods in Molecular Biology. 2022. 2522:87-104.
