Mechanisms of archaeal transcription termination
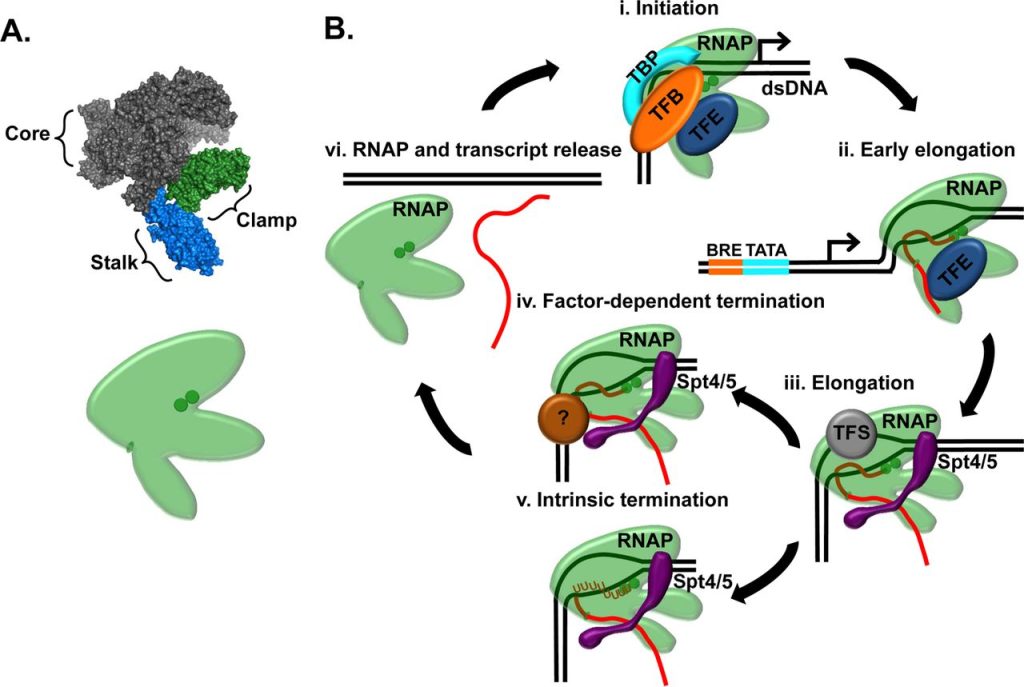
Background
All eukaryotes and most Archaea use histones proteins to bind, wrap and compact their genomes into chromatin. Our understanding of how the transcription apparatus overcomes histone-based barriers and how chromatin structure regulates gene expression is incomplete. Archaeal chromatin and transcription are component simplified but clearly homologous to their eukaryotic counterparts and so provide an attractive, practical experimental system to establish the regulation imposed by histone-bound DNA and the dynamics of such regulation with regards to chromosome structure. Archaeal chromatin is formed with only a single histone protein that wraps DNA to form superhelical structures with the same dimensions as the eukaryotic nucleosome.We will describe the molecular activities of conserved transcription factors that modify RNA polymerase and accelerate transcription through histone-bound DNA.
(1) Establish how histone-based chromatin impacts transcription initiation and elongation and detail the molecular mechanisms of conserved and novel transcription factors that accelerate transcription through histone-bound DNA.
(2) Establish how native histones and histone-variants regulate genomic architecture, RNAP positioning and genome-wide gene expression.
RELEVANT PUBLICATIONS
- Sanders TJ, Ullah F, Gehring AM, Burkhart BW, Vickerman RL, Fernando S, Gardner AF, Ben-Hur A, Santangelo TJ. Extended archaeal histone-based chromatin structure regulates global gene expression in Thermococcus kodakarensis. Frontiers in Microbiology. 2021. May 13; 12:681150.
- Sanders TJ, Marshall CJ, Santangelo TJ. The role of archaeal chromatin in transcription. J Mol Biol. 2019. 431(2):4103-4115.
- Sanders TJ, Lammers M, Marshall C, Walker JE, Lynch ER, Santangelo TJ. TFS and Spt4/5 accelerate transcription through archaeal histone-based chromatin. Mol Micro. 2018, mmi.14191
- Mattiroli F, Bhattacharyya S, Dyer PN, White AE, Sandman K, Burkhart BW, Byrne KR, Lee T, Ahn NG, Santangelo TJ, Reeve JN, Luger K. Structure of histone-based chromatin in Archaea. Science. 2017 Aug 11;357(6351):609-612.
