Mechanisms and control of archaeal DNA repair
Background
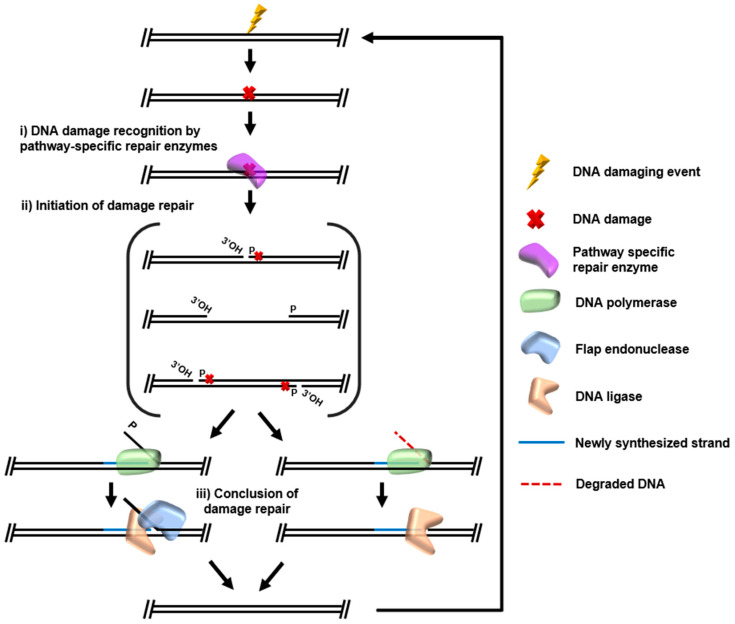
Archaea often thrive in extreme environments that accelerate DNA damage. We lack a comprehensive understanding of the molecular features that impart the extreme resistance to DNA damage exhibited by many Archaea. We aim to resolve the processes that maintain, replicate, and repair the genome of the model archaeon Thermococcus kodakarensis. Our groups have worked together for ~ 15 years and coauthored > 20 manuscripts. The proposed studies expand our collaboration, expose students to academic, industrial and national laboratories, and develop and implement new techniques that will expand studies of DNA replication and repair across Domains. We employ a scaled-approach, from purified enzyme kinetics on the millisecond time scale to whole cell phenotypic assays and growth measurements. Our collaboration provides a platform to study archaeal DNA repair from avenues that would not be possible without the combined expertise of our groups.With complementary in vitro and in vivo assays, we will i) establish the substrate specificities, rates and activities required for base excision repair (BER) and ii) establish the enzymes responsible for transcription-coupled DNA repair (TCR) and likely provide the first mechanistic evidence for archaeal nucleotide excision repair (NER). We will also elucidate the roles of the two DNAPs – Pol B and Pol D – in mediating DNA repair.
RELEVANT PUBLICATIONS
- Marshall CJ, Santangelo TJ. Archaeal DNA Repair Mechanisms. Biomolecules. 2020. Oct 23; 10(11):1472.
- Gehring AM, Zatopek KM, Burkhart BW, Potapov V, Santangelo TJ, Gardner AF. Biochemical reconstitution and genetic characterization of the major oxidative damage base excision DNA repair pathway in Thermococcus kodakarensis. DNA Repair (Amst). 2019. 86:102767.
- Gehring AM, Santangelo TJ. Archaeal RNA polymerase arrests transcription at DNA lesions. Transcription. 2017;8(5):288-296.
- Burkhart BW, Cubonova L, Heider MR, Kelman Z, Reeve JN, Santangelo TJ. The GAN exonuclease or the flap endonuclease and RNase HII are necessary for viability of Thermococcus kodakarensis. J Bacteriol. 2017. 199(13): e00141-17.
- Heider MR, Burkhart BW, Santangelo TJ, Gardner AF. Defining the RNaseH2 enzyme-initiated ribonucleotide excision repair pathway in Archaea. J Biol Chem. 2017. 292(21):8835-8845.
