Establishing the archaeal epitranscriptome
Sas-chen et al., 2020
Background
Maintenance of RNA structures at elevated temperatures – or at the extremes of salinity, pressure or pH – is likely facilitated by epitranscriptome modifications. Our goal is to establish the Rules of mRNA Modifications by exploiting the diverse, abundant, and extensive modifications that are present in archaeal mRNAs. We will use a combination of unbiased techniques and genomic ribonucleoside modification specific mapping techniques to identify, map, and compare the totality of ribonucleoside modifications present in a diverse range of extremophilic Archaea. We will identify and characterize a suite of archaeal enzymes responsible for generating the archaeal epitranscriptome. We will establish the targeting sequences that result in site- and modification-specific alterations of select mRNAs. The rules of mRNA selection and targeting by each enzyme will be iteratively tested in vitro and then in vivo. Given that the mRNA modification machinery is largely conserved, it is likely that the Rules of mRNA Modification and changes to mRNA structure and function due to modifications are also conserved. Through evolutionary comparisons and in silico approaches we will expand the Rules of mRNA Modification derived from our archaeal studies to the other Domains, based on the conservation of RNA modification enzymes, mRNA targets, and targeting sequences/structures. The results obtained – along with the extensive datasets, mapping techniques, mass spectrometry standards, chemical and enzymatic characterizations of RNA modification enzymes – will be documented in a new web-based platform, the mRNA Modification Database. This database will serve the broader RNA community, STEM researchers, and the public.
RELEVANT PUBLICATIONS
- Scott KA, Williams SA, Santangelo TJ. Thermococcus kodakarensis provides a versatile hyperthermophilic archaeal platform for protein expression. Methods in Enzymology. 2021. 659:243-273.
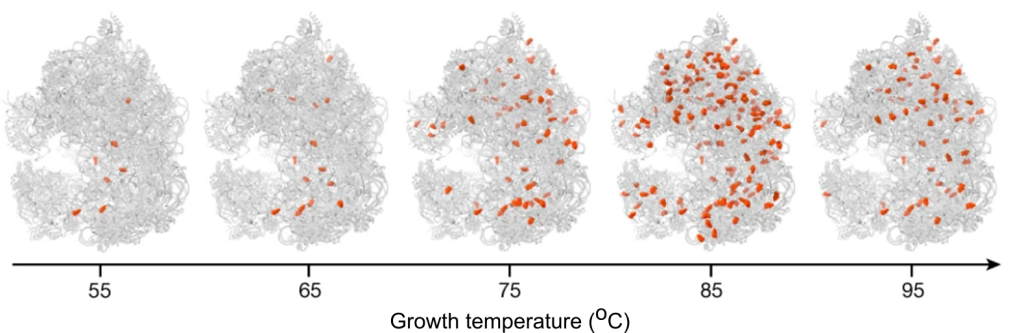
- Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, Gamage ST, Nobe Y, Briney CA, Levy MJ, Fuchs RT, Robb GB, Hartmann J, Sharma S, Lin Q, Florens L, Washburn MP, Isobe T, Santangelo TJ, Shalev-Benami M, Meier JL, Schwartz S. Dynamic RNA acetylation revealed by cross-evolutionary mapping. Nature. 2020. 583(7817):638-643.
- Turner B, Burkhart BW, Widenbach K, Schmitz RA, Ross R, Limbach PA, Draper, DE, de Crecy-Lagard V, Stedman K, Santangelo TJ, Iwata-Reuyl, D. Archaeosine modification of archaeal tRNA – A role in structural stabilization. J Bacteriol, 2020. 202(8):e00748-19.
