Publications
Oct 2024
A novel N4,N4-dymethylcytidine in the Archaeal ribosome enhances hyperthermophily. Fluke KA, Dai N, Wolf EJ, Fuchs RT, Ho PS, Talbott V, Elkins L, Tsai YL, Schiltz J, Febvre HP, Czarny R, Robb GB, Corrêa IR Jr, Santangelo TJ. PNAS. [PubMed]
Sep 2024
Intein splicing efficiency and RadA levels can control the mode of archaeal DNA replication. Liman GLS, Lennon CW, Mandley JL, Galyon AM, Zatopek KM, Gardner AF, Santangelo TJ. Science Advances. [PubMed]
Aug 2024
The extensive m5C epitranscriptome of Thermococcus kodakarensis is generated by a suite of RNA methyltransferases that support thermophily. Fluke KA, Fuchs RT, Tsai YL, Talbott V, Elkins L, Febvre HP, Dai N, Wolf EJ, Burkhart BW, Schiltz J, Brett Robb G, Corrêa IR Jr, Santangelo TJ. Nature comunications. [PubMed]
May 2024
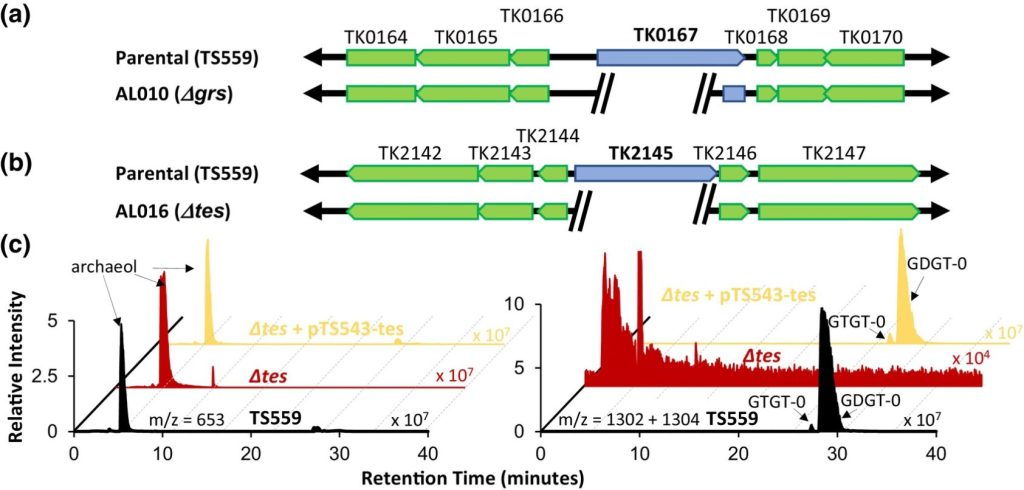
Tetraether archaeal lipids promote long‐term survival in extreme conditions. Liman GLS, Garcia AA, Fluke KA, Anderson HR, Davidson SC, Welander PV, Santangelo TJ. Molecular Microbiology. [PubMed]
Feb 2024
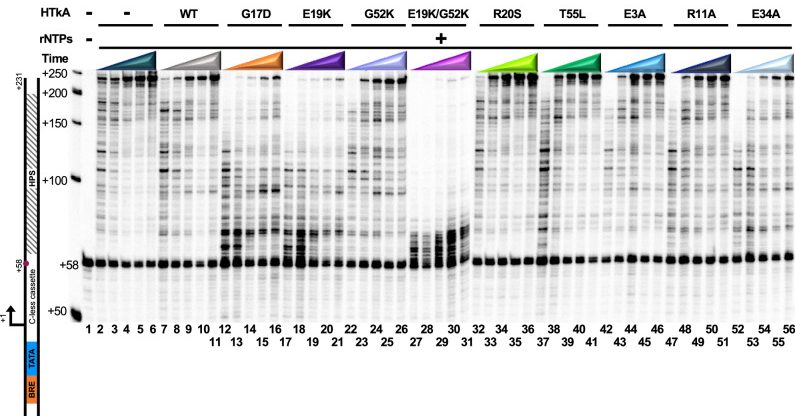
Archaeal histone-based chromatin structures regulate transcription elongation rates. Wenck BR, Vickerman RL, Burkhart BW, Santangelo TJ. Communications Biology. [PubMed]
Aug 2022
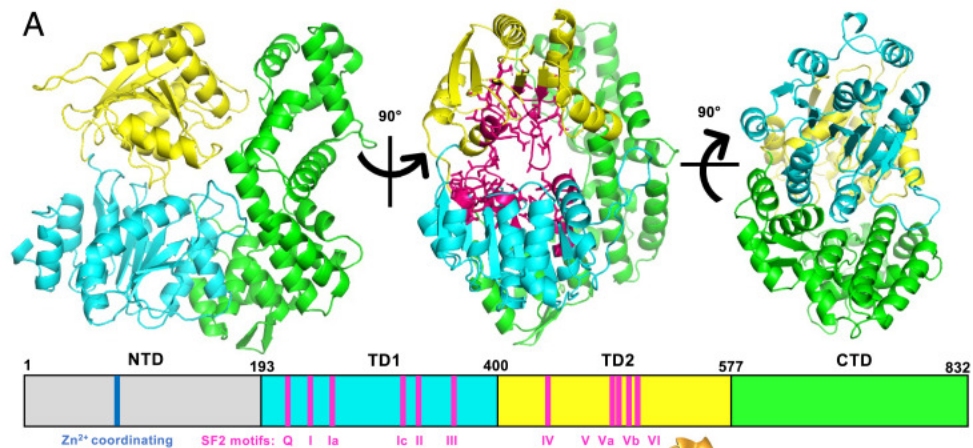
The structure and activities of the archaeal transcription termination factor Eta detail vulnerabilities of the transcription elongation complex. Marshall CJ, Qayyum MZ, Walker JE, Murakami KS, Santangelo TJ. PNAS. [PubMed]
Mar 2022
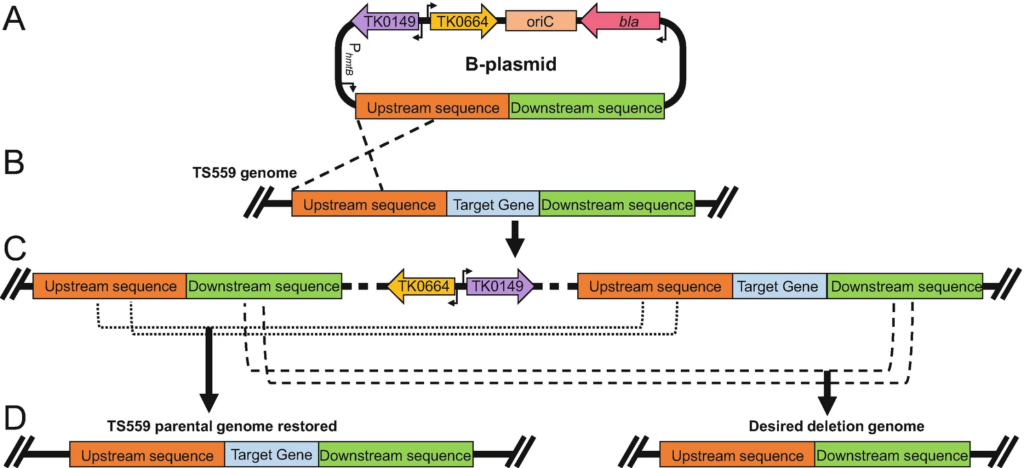
Transformation Techniques for the Anaerobic Hyperthermophile Thermococcus kodakarensis. Liman GLS, Stettler ME, Santangelo TJ. Methods in Molecular Biology. [PubMed]
July 2021
Thermococcus kodakarensis provides a versatile hyperthermophilic archaeal platform for protein expression. Scott KA, Williams SA, Santangelo TJ. Methods in Enzymology. [PubMed]

May 2021
Extended Archaeal Histone-Based Chromatin Structure Regulates Global Gene Expression in Thermococcus kodakarensis. Sanders TJ, Ullah F, Gehring AM, Burkhart BW, Vickerman RL, Fernando S, Gardner AF, Ben-Hur A, Santangelo TJ. Frontier in Microbiology. [PubMed]

Oct 2020
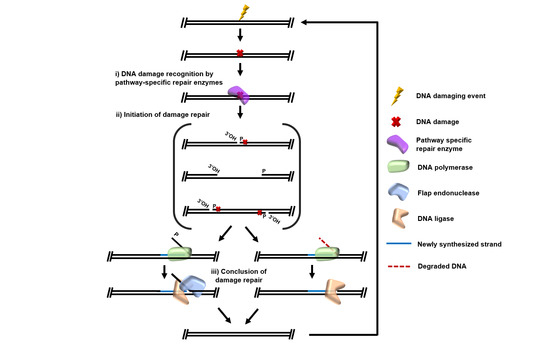
Archaeal DNA Repair Mechanisms. Marshall CJ, Santangelo TJ. Archaeal DNA Repair Mechanisms. Biomolecules. [PubMed]
Apr 2020
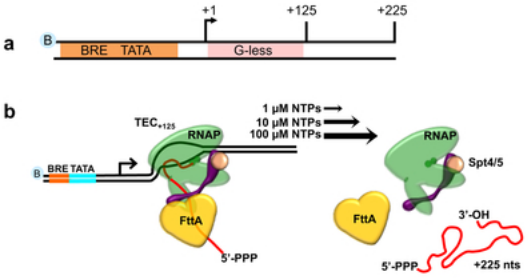
FttA is a CPSF73 homologue that terminates transcription in Archaea. Sanders TJ, Wenck BR, Selan JN, Barker MP, Trimmer SA, Walker JE, Santangelo TJ. Nature Microbiology. [PubMed]
Sep 2019
The Role of Archaeal Chromatin in Transcription. Sanders TJ, Marshall CJ, Santangelo TJ. Journal of Molecular Biology. [PubMed]
Mar 2019
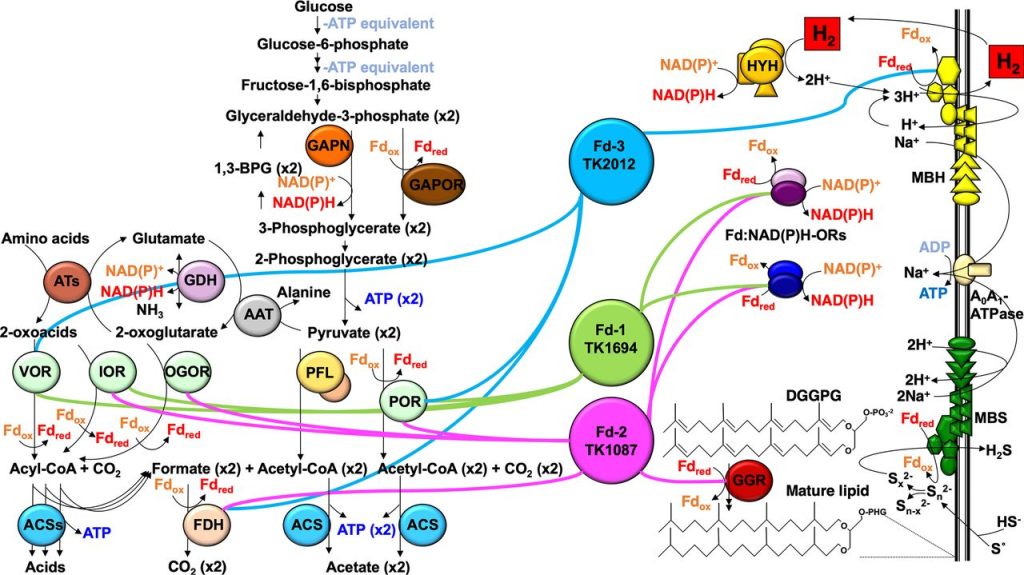
Distinct Physiological Roles of the Three Ferredoxins Encoded in the Hyperthermophilic ArchaeonThermococcus kodakarensis. Burkhart BW, Febvre HP, Santangelo TJ. mBio. [PubMed]
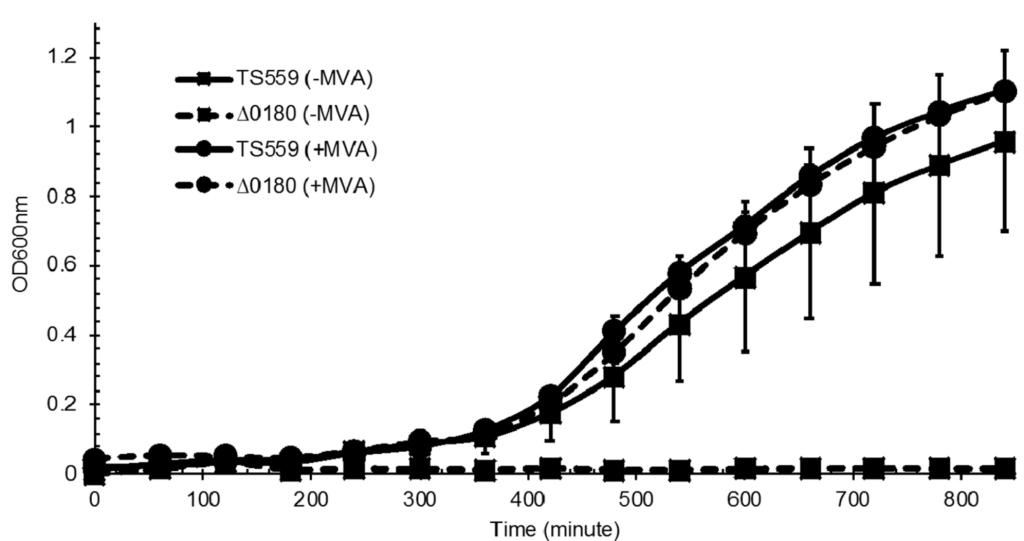
Mar 2019
A linear pathway for mevalonate production supports growth of Thermococcus kodakarensis. Liman GLS, Hulko T, Febvre HP, Brachfeld AC, Santangelo TJ. Extremophiles. [PubMed]
Mar 2019
TFS and Spt4/5 accelerate transcription through archaeal histone-based chromatin. Sanders TJ, Lammers M, Marshall CJ, Walker JE, Lynch ER, Santangelo TJ. Molecular Microbiology. [PubMed]
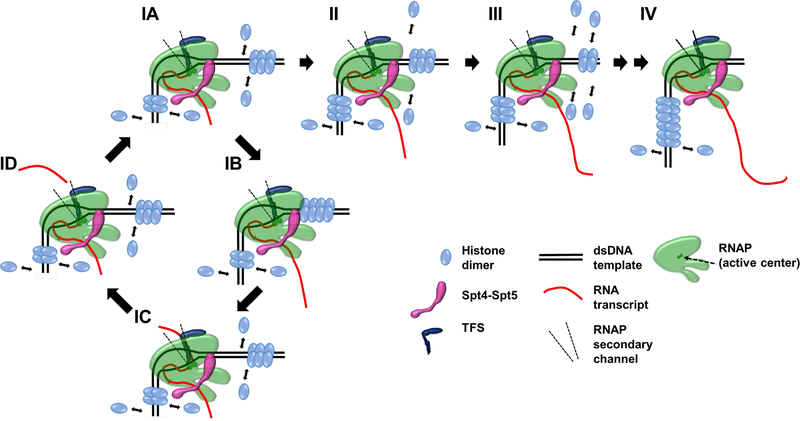
Mar 2018
An Archaeal Fluoride-Responsive Riboswitch Provides an Inducible Expression System for Hyperthermophiles. Speed MC, Burkhart BW, Picking JW, Santangelo TJ. Applied and Environmental Microbiology. [PubMed]
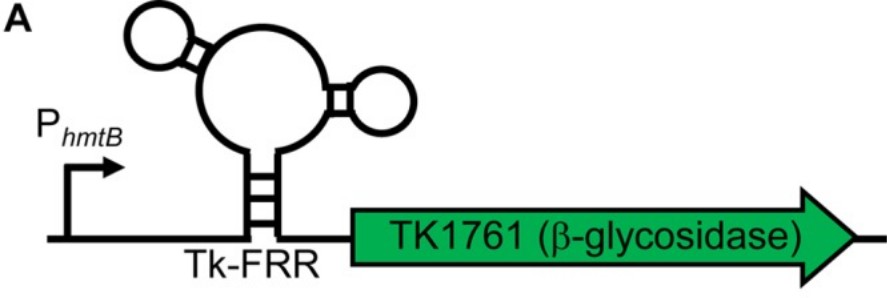
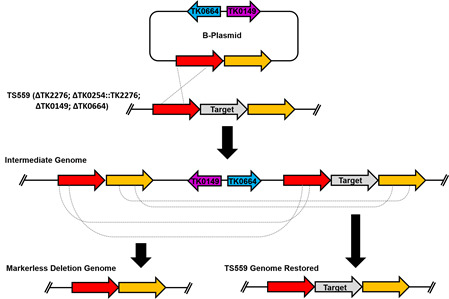
Nov 2017
Markerless Gene Editing in the Hyperthermophilic Archaeon Thermococcus kodakarensis. Gehring AM, Sanders TJ, Santangelo TJ. Biology Protocols. [PubMed]
Oct 2017
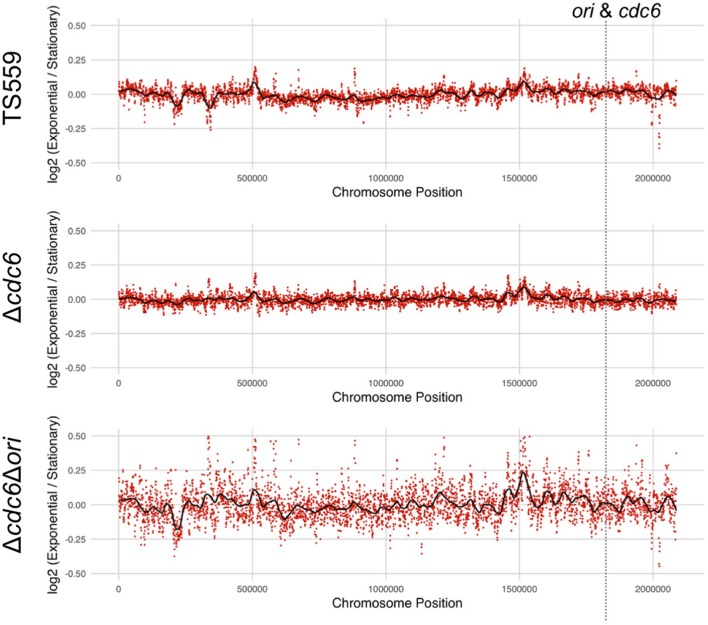
Genome Replication inThermococcus kodakarensis Independent of Cdc6 and an Origin of Replication. Gehring AM, Astling DP, Matsumi R, Burkhart BW, Kelman Z, Reeve JN, Jones KL, Santangelo TJ. Frontiers in MIcrobiology. [PubMed]
Aug 2017
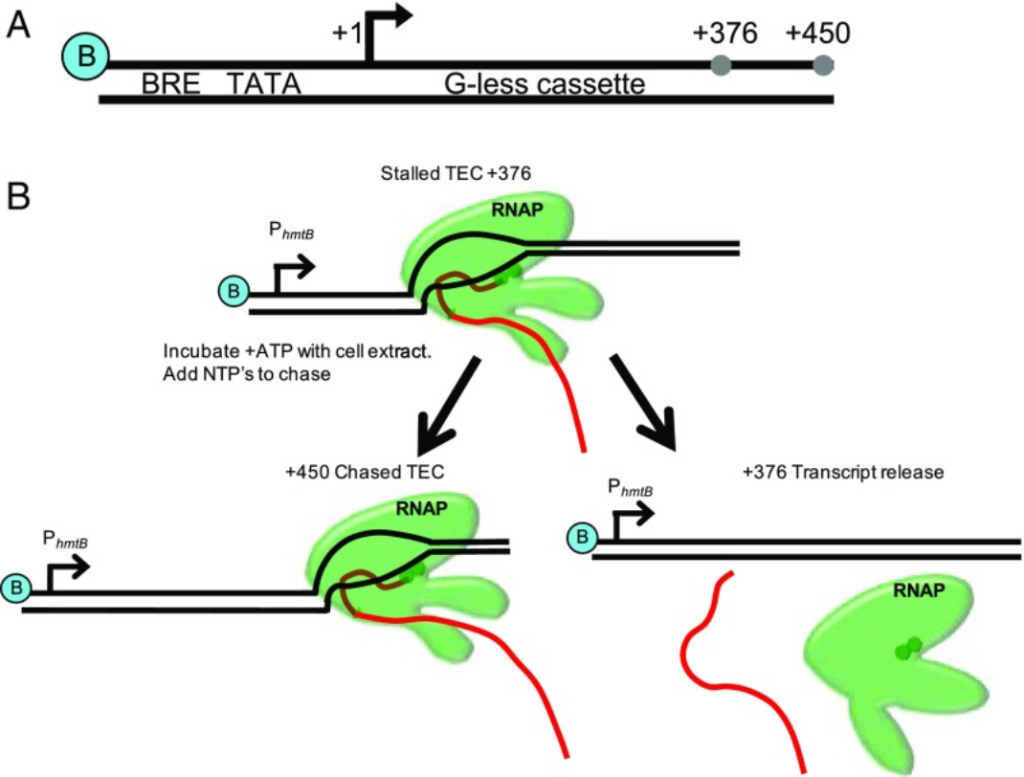
Factor-dependent archaeal transcription termination. Walker JE, Luyties O, Santangelo TJ. PNAS. [PubMed]
June 2017
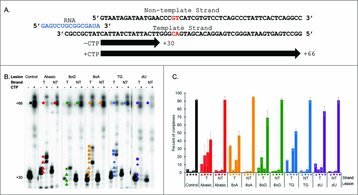
Archaeal RNA polymerase arrests transcription at DNA lesions. Gehring AM, Santangelo TJ. Transcription. [PubMed]
June 2017
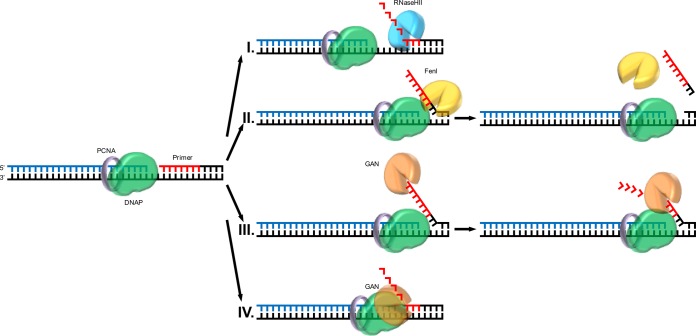
The GAN Exonuclease or the Flap Endonuclease Fen1 and RNase HII Are Necessary for Viability of Thermococcus kodakarensis. Burkhart BW, Cubonova L, Heider MR, Kelman Z, Reeve JN, Santangelo TJ. Journal of Bacteriology. [PubMed]
June 2016
Transcription Regulation in Archaea. Gehring AM, Walker JE, Santangelo TJ. Journal of Bacteriology. [PubMed]
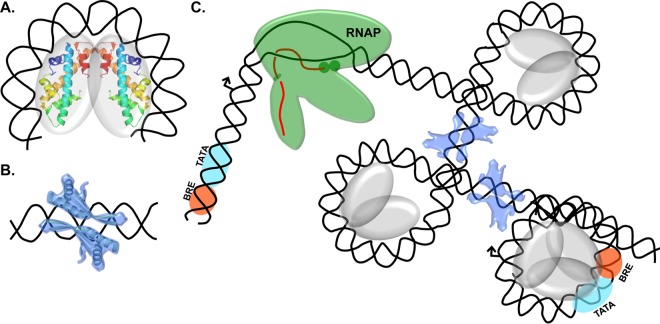
Sep 2015
Analyses of in vivo interactions between transcription factors and the archaeal RNA polymerase. Walker JE, Santangelo TJ.Methods. [PubMed]
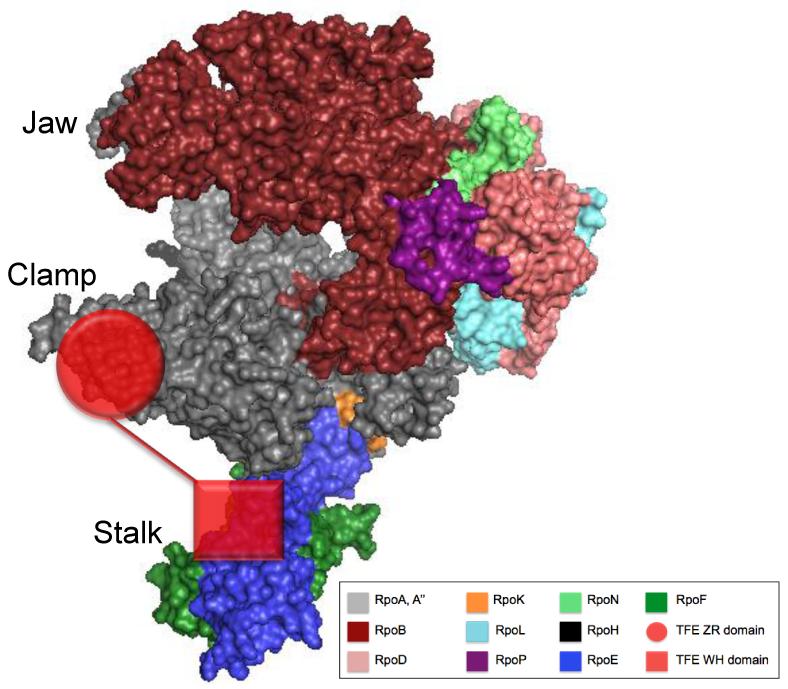
2015
Preface. Bacterial transcriptional control. Artsimovitch I, Santangelo TJ. Methods in Molecular Biology. [PubMed]

2013
Genetic techniques for the archaea. Farkas JA, Picking JW, Santangelo TJ. Annual Reviews in Genetics. [PubMed]
June 2012
Genetics Techniques for Thermococcus kodakarensis. Farkas JA, Hileman TH, Santangelo TJ. Frontiers in MIcrobiology. [PubMed]
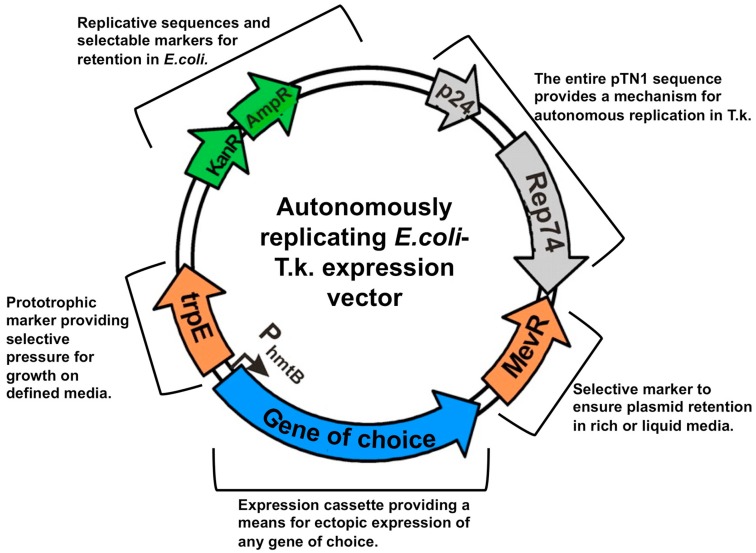
Santangelo et. al.,
Aug 2011
Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Santangelo TJ, Cuboňová L, Reeve JN. Molecular Microbiology. [PubMed]
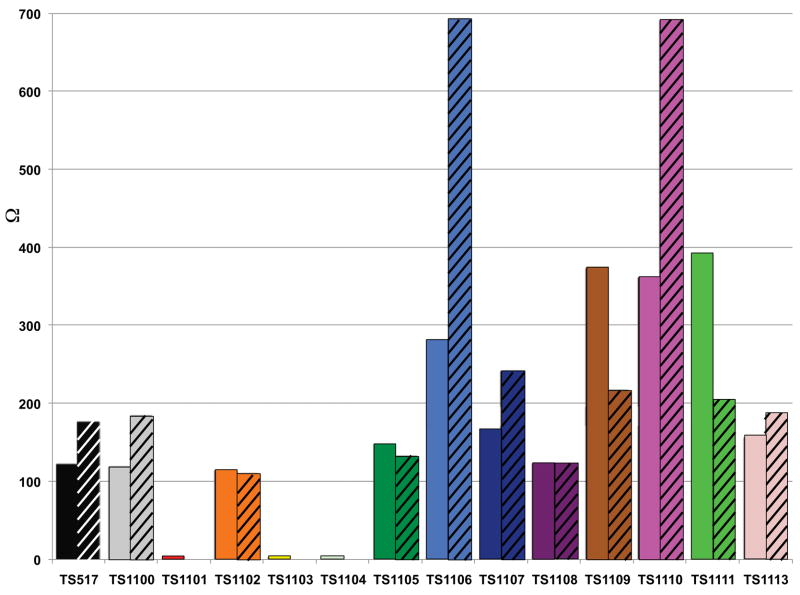
June 2012
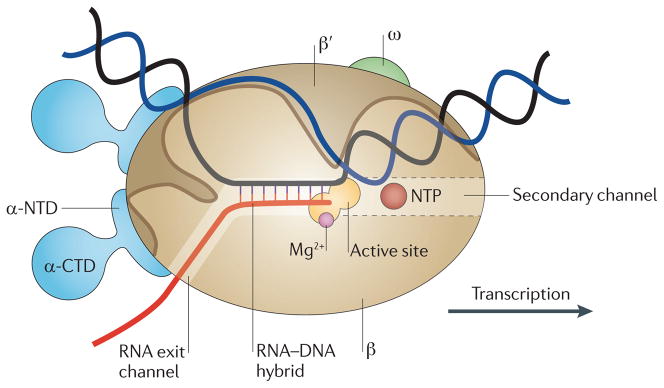
Termination and antitermination: RNA polymerase runs a stop sign. Santangelo TJ, Artsimovitch I. National Reviews in Microbiology. [PubMed]
July 2010
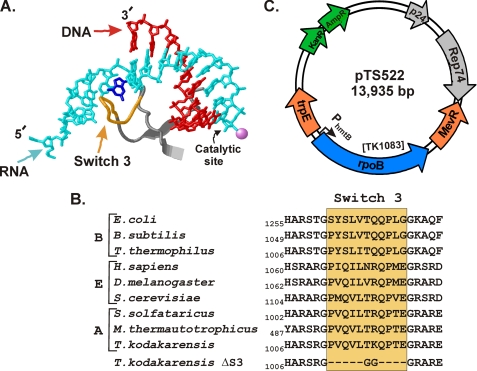
Deletion of switch 3 results in an archaeal RNA polymerase that is defective in transcript elongation. Santangelo TJ, Reeve JN. Journal of Biological Chemistry. [PubMed]
Feb 2010
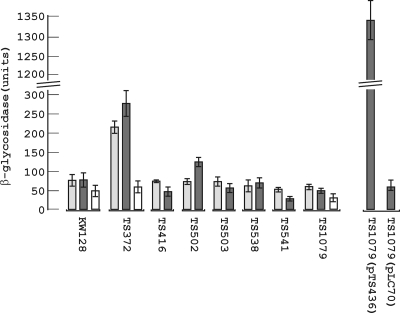
Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Santangelo TJ, Cubonová L, Reeve JN. Applied and Environmental Microbiology. [PubMed]
Nov 2009
Archaeal intrinsic transcription termination in vivo. Santangelo TJ, Cubonová L, Skinner KM, Reeve JN. Journal of Bacteriology. [PubMed]
May 2008
Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Santangelo TJ, Cubonová L, Reeve JN. Applied and Environmental Microbiology. [PubMed]
Mar 2008
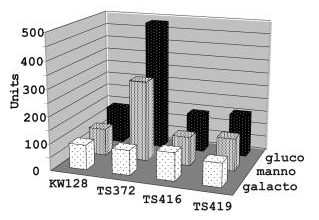
Polarity in archaeal operon transcription in Thermococcus kodakaraensis. Santangelo TJ, Cubonová L, Matsumi R, Atomi H, Imanaka T, Reeve JN. Journal of Bacteriology. [PubMed]
Mar 2007
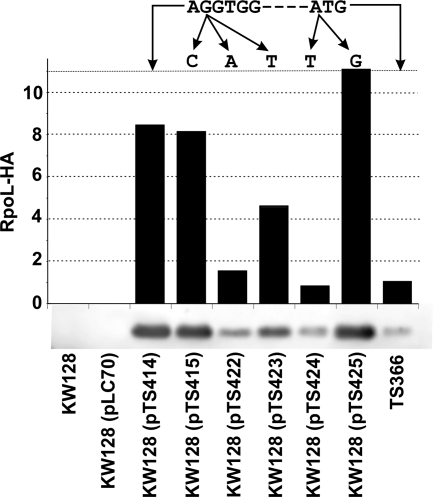
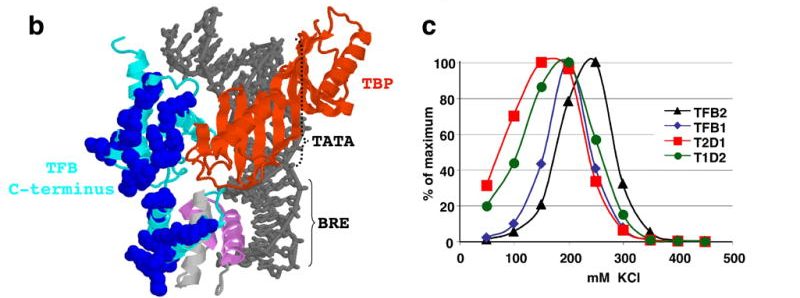
TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. Santangelo TJ, Cubonová L, James CL, Reeve JN. Journal of Bacteriology. [PubMed]
Jan 2006
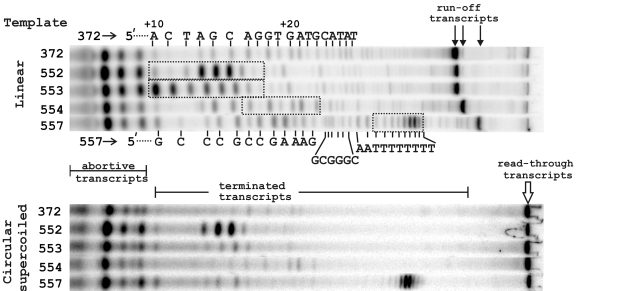
Archaeal RNA polymerase is sensitive to intrinsic termination directed by transcribed and remote sequences. Santangelo TJ, Reeve JN. Journal of Bacteriology. [PubMed]
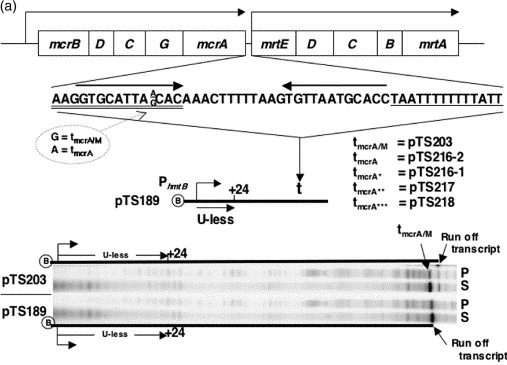
Apr 2004
Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Santangelo TJ, Roberts JW. Molecular Cell. [PubMed]
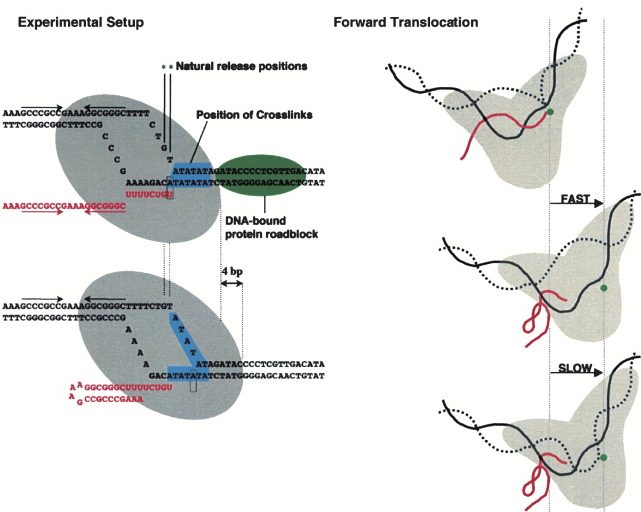
2003
Formation of long DNA templates containing site-specific alkane-disulfide DNA interstrand cross-links for use in transcription reactions. Santangelo TJ, Roberts JW. Methods in Enzymology. [PubMed]
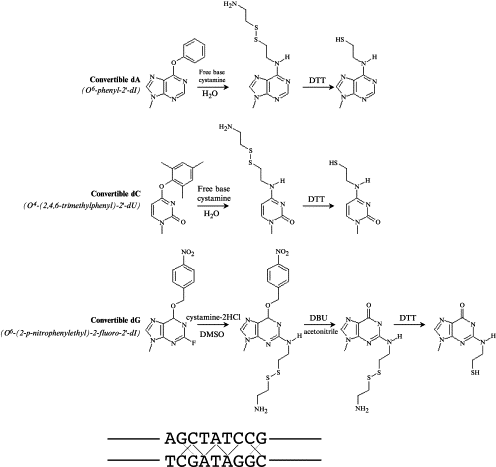
May 2003
RNA polymerase mutations that impair conversion to a termination-resistant complex by Q antiterminator proteins. Santangelo TJ, Mooney RA, Landick R, Roberts JW. Genes and Development. [PubMed]
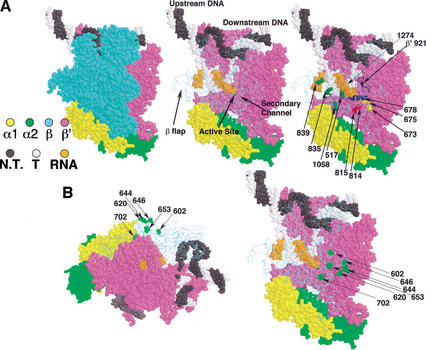
Apr 2002
RfaH, a bacterial transcription antiterminator. Santangelo TJ, Roberts JW. Molecular Cell. [PubMed]
Santangelo Lab
Powered by WordPress